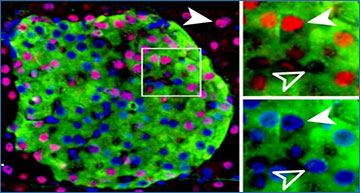
A recently described variety of RNA closely associated with gene expression was found to be largely cell-type specific, raising the possibility this variety of RNA sequences may be able to be used as a marker in stem cell research.
Only about 20 percent of RNA codes for proteins, and the remaining 80 percent, called non-coding RNA, is thought to be involved in a variety of cellular processes including RNA translation, splicing and DNA replication.
Non-coding RNA can be further divided into micro RNA and long non-coding RNA, and over half of long non-coding RNA is chromatin-enriched (cheRNA), where chromatin loops around strands of RNA, which are then bound by RNA polymerase II near the sites of gene promotors, according to previous research. This physical proximity translates to functional connectivity, according to Vasil Galat, PhD, research assistant professor of Pathology at Northwestern University.
In the current study, Galat and the other co-authors of the study discovered that the cheRNA sequences are also specifically associated with different types of cells’ eventual genetic expression.
“Because they are so well associated with a promotor region, they can be used as a predictor of the promotor region’s particular genes,” Galat said. “Once you see the cheRNA expressed, you can judge the location of genes.”
There are several methods biologists currently use to locate gene promotor regions, but this method could be more reliable and precise, according to Galat. Now, the discovery that cheRNA is cell-type specific has particularly tantalizing applications in his primary line of research: pluripotent stem cells.
HIDALGO couples an enhancer at its TSS to the HBG1 promoter
(a) 3C in ASO-transfected K562 cells after sorting reveals a HIDALGO-sensitive contact between the HBG1 promoter and a region near the HIDALGOtranscript #4 TSS. Line arcs depict average relative contact frequency of the two vertices, with s.d. indicated in lighter color. (b) ICeChIP-qPCR quantification of the levels of the three indicated histone lysine methyl marks at HIDALGO TSS #4 and the HBG– promoter, comparing sgRNA3- and ASO1- mediated HIDALGO perturbations to control (‘–’ sgRNA). Error bars represent s.e.m. from three qPCR technical replicates; each ICeChIP was performed on material from a single cell culture. (c) Enhancer assay by relative luciferase activity of the HIDALGO TSS#4 (HID TSS) in K562 cells. HID TSS firefly luciferase activity normalized to co-transfected Renilla luciferase vector. Luciferase (n = 6 independent experiments; error bars represent s.e.m., ****P < 10−4, Welch’s two-tailed t test). (d) Model of HIDALGO activation of HBG1 by bridging the enhancer (HID TSS) to the promoter of HBG1.
Pluripotent stem cells are undifferentiated, meaning they could develop into almost any type of cell in the human body. It can be tricky to keep them in the pluripotent stage, which is why Galat’s lab was invited to collaborate with this University of Chicago-led project.
“Our lab has a great deal of experience working with pluripotent cells,” Galat said. “These cells require experience to maintain. They are spontaneously differentiating all the time in culture.”
Theoretically, by establishing a database of cell types and associated cheRNAs, cheRNA could be used as a marker to narrow down the type of cell a pluripotent stem cell is transforming into — a difficult task with current equipment.
“Many types of cells all look very similar in culture,” Galat said. “You can check markers, but many markers overlap — so to distinguish cell type you have to use many markers. Instead, if you can isolate the cheRNA, it could define the kind of cells you’re dealing with and how functionally mature they are.”
He even hopes scientists could use cheRNA to actively direct differentiation, at some point in the future.
More precise manipulation of pluripotent cells could hasten the process of genetic engineering cells with a specific function — for example, creating highly functional cells of the immune system, which are involved in almost every aspect of health.
“That’s the most interesting feature of cheRNA,” Galat said. “It could serve as a way of characterizing cell type, but also as a method to direct a pluripotent cell to develop into a particular cell type.”
Source – Northwestern University
 lncRNA Blog lncRNA Research and Industry News
lncRNA Blog lncRNA Research and Industry News