Large noncoding RNAs are emerging as an important component in cellular regulation. Considerable evidence indicates that these transcripts act directly as functional RNAs rather than through an encoded protein product. However, a recent study of ribosome occupancy reported that many large intergenic ncRNAs (lincRNAs) are bound by ribosomes, raising the possibility that they are translated into proteins. Here, researchers from ...
Read More »LncRNA: Why and How to Study it?
Introduction Long non-coding RNAs (LncRNAs) are evolutionarily conserved, longer than 200nt, non-coding RNA molecules found in eukaryotes. Growing evidence suggests that LncRNAs have emerged as important regulators for diverse functions1. LncRNAs are involved in a surprisingly wide variety of cellular functions, including epigenetic silencing, transcriptional regulation, RNA processing, and RNA modification2. In addition, LncRNAs have been associated with human diseases ...
Read More »Featured long non-coding RNA – nuclear-enriched abundant transcript 1_2 (NEAT1_2)
A long non-coding RNA (lncRNA), nuclear-enriched abundant transcript 1_2 (NEAT1_2), constitutes nuclear bodies known as “paraspeckles”. Mutations of RNA binding proteins, including TAR DNA-binding protein-43 (TDP-43) and fused in sarcoma/translocated in liposarcoma (FUS/TLS), have been described in amyotrophic lateral sclerosis (ALS). ALS is a devastating motor neuron disease, which progresses rapidly to a total loss of upper and lower motor ...
Read More »Shedding Light on Unique RNAs
from Bioscience Technology The genes that code for proteins—more than 20,000 in total—make up only about 1 percent of the complete human genome. That entire thing—not just the genes, but also genetic junk and all the rest—is coiled and folded up in any number of ways within the nucleus of each of our cells. Think, then, of the challenge that ...
Read More »Seminar – Measurement and analysis of mRNAs and long non-coding RNAs using RNA-Seq
H3ABioNet – Pan African Bioinformatics Network for H3Africa These one day courses are designed for scientists and clinicians with little or no experience in next generation sequencing. The courses aim to provide the experimental and bioinformatics skills required to prepare samples, quantify the levels of mRNA/long-non-coding RNA (RNAseq) or exon/DNA (DNAseq) expresion. We assume that sequencing will be performed by ...
Read More »DETR'PROK: detection of ncRNAs in prokaryotes
RNA-seq experiments are now routinely used for the large scale sequencing of transcripts. In bacteria or archaea, such deep sequencing experiments typically produce 10-50million fragments that cover most of the genome, including intergenic regions. In this context, the precise delineation of the non-coding elements is challenging. Non-coding elements include untranslated regions (UTRs) of mRNAs, independent small RNA genes (sRNAs) and ...
Read More »Pervasive Transcription of the Human Genome Produces Thousands of Previously Unidentified Long Intergenic Noncoding RNAs
Known protein coding gene exons compose less than 3% of the human genome. The remaining 97% is largely uncharted territory, with only a small fraction characterized. The recent observation of transcription in this intergenic territory has stimulated debate about the extent of intergenic transcription and whether these intergenic RNAs are functional. Here, researchers at the University of California, San Francisco ...
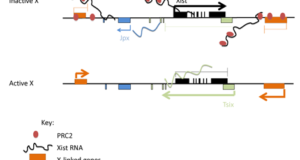
Read More »Guided by RNAs: X-inactivation as a model for lncRNA function
The recent revolution in sequencing technology has helped to reveal a large transcriptome of long non-coding RNAs (lncRNAs). A major challenge in the years to come is to determine what biological functions, if any, they serve. Although the purpose of these transcripts is largely unknown at present, existing examples suggest that lncRNAs play roles in a wide variety of biological ...
Read More »Featured long non-coding RNA – Kcna2 antisense RNA
Neuropathic pain is a refractory disease characterized by maladaptive changes in gene transcription and translation in the sensory pathway. Long noncoding RNAs (lncRNAs) are emerging as new players in gene regulation, but how lncRNAs operate in the development of neuropathic pain is unclear. Here, researchers from the Johns Hopkins University School of Medicine identify a conserved lncRNA, named Kcna2 antisense ...
Read More »Science Gone Social
by Tracy Vence at Genetic Engineering News.com From keeping up with the literature to sparking collaborations and finding funds, scientists are storming social media. Not many scientists have produced manuscripts as a direct result of participating in discussions on social media. Fewer still support their research programs in full by funds obtained in the same way. But, respectively speaking, Emily ...
Read More » lncRNA Blog lncRNA Research and Industry News
lncRNA Blog lncRNA Research and Industry News