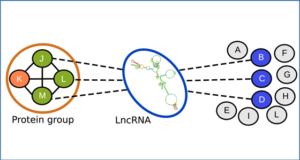
The human transcriptome contains thousands of long non-coding RNAs (lncRNAs). Characterizing their function is a current challenge. An emerging concept is that lncRNAs serve as protein scaffolds, forming ribonucleoproteins and bringing proteins in proximity. However, only few scaffolding lncRNAs have been characterized and the prevalence of this function is unknown. Here, researchers from the Aix-Marseille University, Inserm propose the first computational ...
Read More »FEELnc – a tool for long non-coding RNA annotation
Whole transcriptome sequencing (RNA-seq) has become a standard for cataloguing and monitoring RNA populations. One of the main bottlenecks, however, is to correctly identify the different classes of RNAs among the plethora of reconstructed transcripts, particularly those that will be translated (mRNAs) from the class of long non-coding RNAs (lncRNAs). Here, researchers from University Rennes1 present FEELnc (FlExible Extraction of ...
Read More »Call for papers – non-coding RNA methods
The journal Non-Coding RNA is now accepting submissions for a special issue on Non Coding RNA methods. The special issue co guest-edited by Dr. Piero Carninci from the RIKEN Center for Life Science Technologies in Japan and Dr. Florent Hubé from the National Center for Scientific Research (CNRS) in Paris. This special issue will group together manuscripts that report on ...
Read More » lncRNA Blog lncRNA Research and Industry News
lncRNA Blog lncRNA Research and Industry News