Large noncoding RNAs can localize to regulatory DNA targets by exploiting the three-dimensional architecture of the genome. Jesse Engreitz Harvard-MIT Division of Health Sciences and Technology Massachusetts Institute of Technology Talk at Biology of Genomes 2013 Cold Spring Harbor, NY May 8, 2013
Read More »Nuclear structure, paraspeckles and long non-coding RNA
Walter and Eliza Hall Institute Postgraduate lecture 3 June 2013 Dr Archa Fox WA Institute for Medical Research
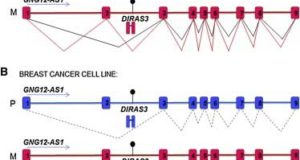
Read More »Imprinted Chromatin around DIRAS3 Regulates Alternative Splicing of GNG12-AS1, a Long Noncoding RNA
Imprinted gene clusters are regulated by long noncoding RNAs (lncRNAs), CCCTC binding factor (CTCF)-mediated boundaries, and DNA methylation. DIRAS3 (also known as ARH1 or NOEY1) is an imprinted gene encoding a protein belonging to the RAS superfamily of GTPases and is located within an intron of a lncRNA called GNG12-AS1. In this study, researchers at Cancer Research UK investigated whether ...
Read More »Differential Expression of Long Non-Coding RNAs in the Livers of Female B6C3F1 Mice
The mammalian genome is transcribed into mRNAs that code for protein as well as a broad spectrum of other noncoding (nc) RNA products. Long ncRNAs (lncRNA), defined as ncRNA species > 200 nucleotides long, are emerging as important epigenetic regulators of gene expression that are involved in a spectrum of biological processes of relevance to toxicology. A team led by ...
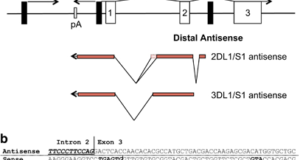
Read More »Featured long non-coding RNA: KIR antisense
Human NK cells express cell surface class I MHC receptors (killer cell immunoglobulin-like receptor, KIR) in a probabilistic manner. Previous studies have shown that a distal promoter acts in conjunction with a proximal bidirectional promoter to control the selective activation of KIR genes. Researchers at SAIC-Frederick report here the presence of an intron 2 promoter in several KIR genes that ...
Read More »Webinar: Long non-coding RNAs in Development & Disease Progression
Long noncoding RNAs (LncRNAs) are eukaryotic RNA molecules greater than 200 nucleotides in length that have no protein-coding capacity. Recently, LncRNAs have emerged as important factors in gene regulation. The inappropriate expression of some LncRNAs has been demonstrated to cause diseases such as liver cancer and Alzheimer’s. Ed Davis, Ph.D., Technical Support Specialist at Arraystar, Inc. discusses the biological and ...
Read More »Webinar: Detection of Long Noncoding RNAs by Stellaris FISH Probes
Long non-coding RNAs are now appearing from the genome’s dark matter. Much of what was previously labeled as junk DNA in fact produces non-coding RNA. While non-coding RNAs serve important biological functions, the lack of protein products requires new tools for detection and quantification. To meet this need, Stellaris RNA FISH probes from Biosearch Technologies are now entering laboratories around ...
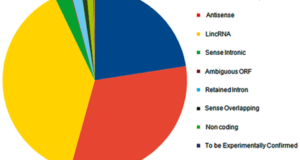
Read More »lncRNome: a comprehensive knowledgebase of human long noncoding RNAs
The advent of high-throughput genome scale technologies has enabled us to unravel a large amount of the previously unknown transcriptionally active regions of the genome. Recent genome-wide studies have provided annotations of a large repertoire of various classes of noncoding transcripts. Long noncoding RNAs (lncRNAs) form a major proportion of these novel annotated noncoding transcripts, and presently known to be ...
Read More »Jpx RNA Activates Xist by Evicting CTCF
In mammals, dosage compensation between XX and XY individuals occurs through X chromosome inactivation (XCI). The noncoding Xist RNA is expressed and initiates XCI only when more than one X chromosome is present. Current models invoke a dependency on the X-to-autosome ratio (X:A), but molecular factors remain poorly defined. Here, researchers at the Howard Hughes Medical Institute demonstrate that molecular ...
Read More »Long Noncoding RNA: a New Player of Heart Failure?
One the most important discoveries of the post-genomic era is that a large fraction of the genome transcribes a heterogeneous population of noncoding RNAs (ncRNA). ncRNAs shorter than 200 nucleotides are usually identified as short/small ncRNAs-examples include PIWI-interacting RNAs, small interfering RNAs, and microRNAs (miRNAs)-whereas those longer than 200 nucleotides are classified as long ncRNAs (lncRNAs). These molecules are emerging ...
Read More » lncRNA Blog lncRNA Research and Industry News
lncRNA Blog lncRNA Research and Industry News