Colorectal cancer (CRC) was one of the most commonly diagnosed malignancies. The molecular mechanisms involved in the progression of CRC remain unclear. Accumulating evidences showed that long noncoding RNAs (lncRNAs) played key roles in tumorigenesis, cancer progression, and metastasis. Researchers at Fudan University aimed to explore the roles of lncRNAs in the progression of CRC.
In this study, the researchers aimed to identify differentially expressed lncRNAs and messenger RNAs (mRNAs) in CRC by analyzing a cohort of previously published datasets: GSE64857. GO and KEGG pathway analyses were applied to give an insight in the functions of those lncRNAs and mRNAs in CRC.
Totally, 46 lncRNAs were identified as differentially expressed between stage II and stage III CRC for the first time screening by microarray. GO and KEGG pathway analyses showed that differentially expressed lncRNAs were involved in regulating signal transduction, cell adhesion, cell differentiation, focal adhesion, and cell adhesion molecules.
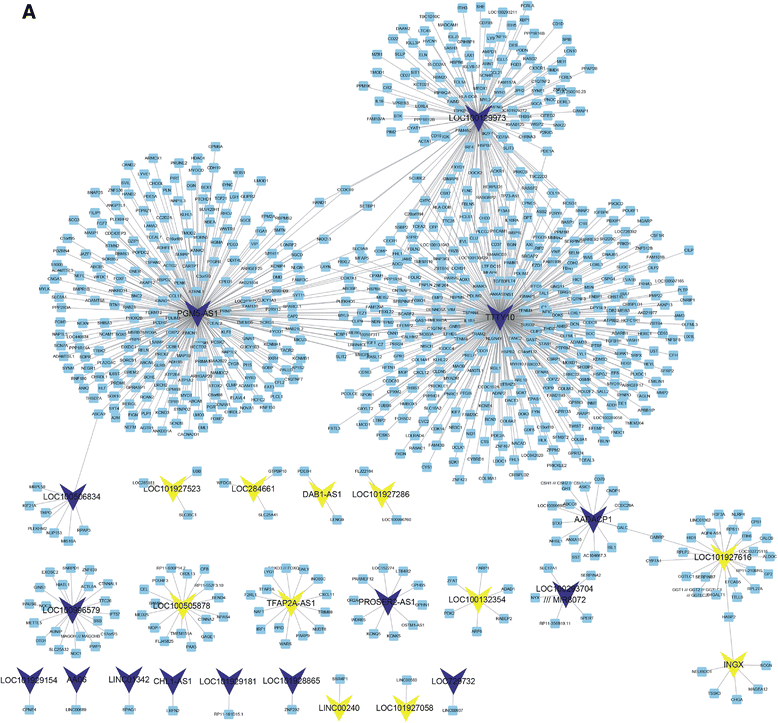
Construction of co-expressed DEG-lncRNA networks in colorectal cancer between stage II and stage III. a Totally, 46 lncRNAs and 881 differentially expressed genes were clustered in the network
The researchers found three lncRNAs (LOC100129973, PGM5-AS1, and TTTY10) widely co-expressed with differentially expressed mRNAs. They also constructed lncRNA-associated PPI in CRC and found that these lncRNAs may be associated with CRC progression. Moreover, they found that high PGM5-AS1 expression levels were associated with worse overall survival in CRC cancer. The researchers believe that this study would provide novel potential therapeutic and prognostic targets for CRC.
 lncRNA Blog lncRNA Research and Industry News
lncRNA Blog lncRNA Research and Industry News