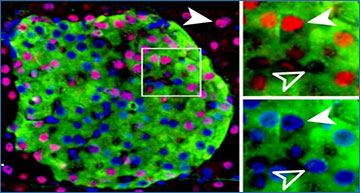
Long intergenic noncoding RNAs (lincRNAs) have emerged as key regulators of cellular functions and physiology. Yet functional lincRNAs often have low, context-specific and tissue-specific expression. Researchers from Columbia University Medical Center hypothesized that many human monocyte and adipose lincRNAs would be absent in current public annotations due to lincRNA tissue specificity, modest sequencing depth in public data, limitations of transcriptome assembly algorithms and lack of dynamic physiological contexts. Deep RNA-seq was performed in peripheral blood CD14+ monocytes (monocytes; average ~247 million reads per sample) and adipose tissue (average ~378 million reads per sample) collected before and after human experimental endotoxemia, an in vivo inflammatory stress, to identify tissue-specific and clinically relevant lincRNAs. Using a stringent filtering pipeline, the researchers identified 109 unannotated lincRNAs in monocytes and 270 unannotated lincRNAs in adipose. Most unannotated lincRNAs are not conserved in rodents and are tissue-specific while many have features of regulated expression and are enriched in transposable elements. Specific subsets have enhancer RNA characteristics or are expressed only during inflammatory stress. A subset of unannotated lincRNAs were validated and replicated for their presence and inflammatory induction in independent human samples and for their monocyte and adipocyte origins. Through interrogation of public genome-wide association data, they also found evidence of specific disease association for selective unannotated lincRNAs. These findings highlight the critical need to perform deep RNA-seq in a cell-, tissue- and context- specific manner in order to annotate the full repertoire of human lincRNAs for a complete understanding of lincRNA roles in dynamic cell functions and in human disease.
Trait-associated SNPs overlap unannotated lincRNA CUFF.199812 at GWAS loci
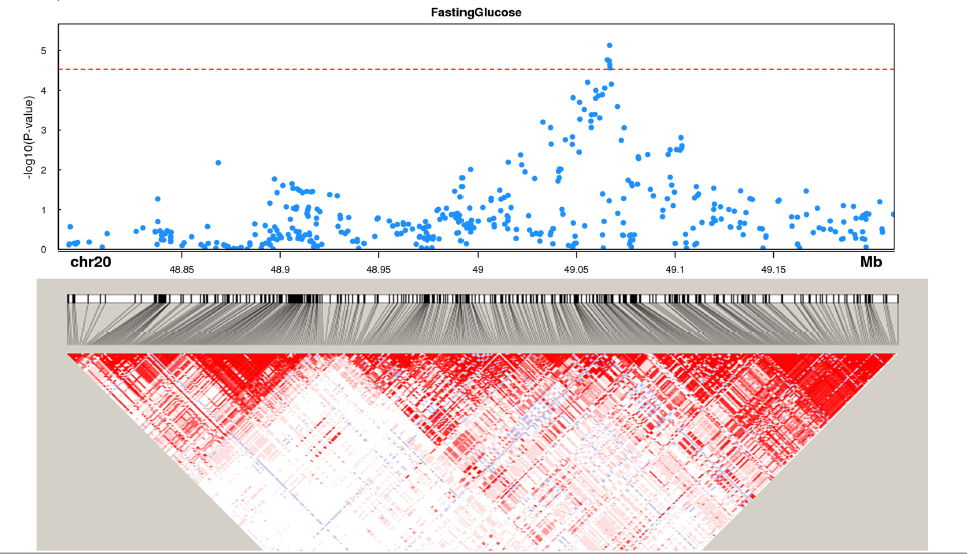
Gene annotation, trait-associated SNPs and LD structure are shown for CUFF.199812. Genes from GENCODE v19 are plotted in each locus. Protein coding genes are shown in blue, unannotated lincRNAs in red, other lincRNAs in green. All other genes, e.g., COX6CP2, are shown in black. P-value cutoff is corrected for the number of SNPs mapped to lincRNAs. LD heatmap color scheme is the default D’/LOD scheme in Haploview.
 lncRNA Blog lncRNA Research and Industry News
lncRNA Blog lncRNA Research and Industry News