A new class of transcripts, long non-coding RNAs (lncRNAs), has been recently found to be pervasively transcribed in the genome. These mRNA-like molecules, which lack significant protein-coding capacity, once thought to be a part of the dark matter, now have been implicated in a wide range of biological functions through diverse and as yet poorly understood molecular mechanisms. Multiple facets ...
Read More »Role of microRNAs and long-non-coding RNAs in CD4(+) T-cell differentiation
CD4(+) T lymphocytes orchestrate adaptive immune responses by differentiating into various subsets of effector T cells such as T-helper 1 (Th1), Th2, Th17, and regulatory T cells. These subsets have been generally described by master transcription factors that dictate the expression of cytokines and receptors, which ultimately define lymphocyte effector functions. However, the view of T-lymphocyte subsets as stable and ...
Read More »Long noncoding RNAs in development and disease of the central nervous system
The central nervous system (CNS) is a complex biological system composed of numerous cell types working in concert. The intricate development and functioning of this highly ordered structure depends upon exquisite spatial and temporal control of gene expression in the cells comprising the CNS. Thus, gene regulatory networks that control cell fates and functions play critical roles in the CNS. ...
Read More »RNA: The genome's rising stars
from Nature by Amy Maxmen When Saba Valadkhan lingered in the hallways at conferences, absorbed in discussions about the strands of ‘junk’ DNA that litter the human genome, she was not looking for work. She was consumed with curiosity about the possibility that long RNA sequences that do not encode proteins nevertheless have a function — enhancing or suppressing gene ...
Read More »Predicting long non-coding RNAs using RNA sequencing
The advent of next-generation sequencing, and in particular RNA-sequencing (RNA-Seq), technologies has expanded our knowledge of the transcriptional capacity of human and other animal, genomes. In particular, recent RNA-Seq studies have revealed that transcription is widespread across the mammalian genome, resulting in a large increase in the number of putative transcripts from both within, and intervening between, known protein-coding genes. ...
Read More »MALAT1 – a paradigm for long noncoding RNA function in cancer
The metastasis-associated lung adenocarcinoma transcript 1 (MALAT1) is a bona fide long noncoding RNA (lncRNA). MALAT1, also known as nuclear-enriched transcript 2 (NEAT2), was discovered as a prognostic marker for lung cancer metastasis but also has been linked to several other human tumor entities. Recent work established a critical regulatory function of this lncRNA in lung cancer metastasis and cell ...
Read More »X-Inactivation, Imprinting, and Long Noncoding RNAs in Health and Disease
X chromosome inactivation and genomic imprinting are classic epigenetic processes that cause disease when not appropriately regulated in mammals. Whereas X chromosome inactivation evolved to solve the problem of gene dosage, the purpose of genomic imprinting remains controversial. Nevertheless, the two phenomena are united by allelic control of large gene clusters, such that only one copy of a gene is ...
Read More »miRNAs and long noncoding RNAs as biomarkers in human diseases
Noncoding RNAs (ncRNAs) are transcripts that have no apparent protein-coding capacity; however, many ncRNAs have been found to play a major biological role in human physiology. Their deregulation is implicated in many human diseases, but their exact roles are only beginning to be elucidated. Nevertheless, ncRNAs are extensively studied as a novel source of biomarkers, and the fact that they ...
Read More »Long Noncoding RNAs – Cellular Address Codes in Development and Disease
In biology as in real estate, location is a cardinal organizational principle that dictates the accessibility and flow of informational traffic. An essential question in nuclear organization is the nature of the address code—how objects are placed and later searched for and retrieved. Long noncoding RNAs (lncRNAs) have emerged as key components of the address code, allowing protein complexes, genes, ...
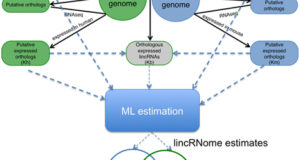
Read More »Twice as many human and mouse lincRNAs than protein-coding genes
Genome analysis of humans and other mammals reveals a surprisingly small number of protein-coding genes, only slightly over 20,000 (although the diversity of actual proteins is substantially augmented by alternative transcription and alternative splicing). Recent analysis of the mammalian genomes and transcriptomes, in particular, using the RNAseq technology, shows that, in addition to protein-coding genes, mammalian genomes encode many long ...
Read More » lncRNA Blog lncRNA Research and Industry News
lncRNA Blog lncRNA Research and Industry News